![]() In the recent PRTools updates of September 2012 (4.2.2) November 2012 (4.2.3) and January 2013 (4.2.4) a number of tools have been added and changed of which not everybody might be aware. Here we will pay more attention to them and give some background information about their use. It concerns some new classifiers, the handling of doubles where originally datasets where expected, the mapping overload of dyadic operations, some routines that facilitate the programming of mappings, batch processing, the order of mapping output parameters and some minor issues.
In the recent PRTools updates of September 2012 (4.2.2) November 2012 (4.2.3) and January 2013 (4.2.4) a number of tools have been added and changed of which not everybody might be aware. Here we will pay more attention to them and give some background information about their use. It concerns some new classifiers, the handling of doubles where originally datasets where expected, the mapping overload of dyadic operations, some routines that facilitate the programming of mappings, batch processing, the order of mapping output parameters and some minor issues.
Classifiers
For some researchers pattern recognition is classification. Readers of this blog will have noticed that we see this somewhat different. Representation is the key issue of pattern recognition. However, the tool set should contain a set of good and preferably very different classifiers, in spite of David Hand’s observation that the progress in classifier technology might be an illusion. Anyway, we are glad that every now and then PRTools users submit a new classifier. Here are some recent examples.
Many have already wondered where the PRTools version is of Breiman’s famous proposal of the random forest: a randomized collection of small decision trees based on different feature subsets. Well, recently David Tax created a fully Matlab based version of this proposal, randomforestc. It has a good performance but is slow. May be a mex-file based version will become available later. In a future post on classifier benchmarking will be discussed what a good classifier performance means.
 One of the first classifiers in PRTools is the decision tree classifier
One of the first classifiers in PRTools is the decision tree classifier treec. It was created by some students who studied Breiman’s (again!) classic: Classification and regression trees, (1984), CART. Later it was extended with some pruning algorithms proposed by Quinlan and Chou. Quinlan is the author of the well known C4.5 decision tree package. His papers and proposals have been studied by Sergey Verzakov, who created an upgrade of treec, called dtc. In a number of experiments it appeared to have a performance close to randomforestc. It is, however, much faster as it is based on a single tree. We hope that Sergey in the near future can extend the Verzakov Tree with some visualization tools.
Neural networks still constitute an inspiring paradigm for classifier design. There is a strong relation with the field of combining classifiers in case the layers have a different architecture and/or the training of the layers is done consecutively. Laurens van der Maaten, the author of the dimensionality reduction toolbox, contributed with two classifiers. The first, the Voted Perceptron (vpc) is based on a paper by Freund and Schapire and combines an ensemble of perceptrons by voting. The second one follows a proposal by Larochelle and Bengio on the Discriminative Restricted Boltzmann Machine (RBM) (drbmc). The model introduces binary stochastic latent variables in logistic regression, which turns it into a powerful non-linear model. The stochastic latent variables act a lot like rectified linear units in neural networks. For both proposals holds that they are much faster than available neural network classifiers bpxnc and lmnc. RBM may perform very well.
Doubles instead of datasets
When PRTools was designed it was decided that objects should be represented by datasets: the feature vectors, labels and all annotation in a single variable. All routines were based on that and tests were included to check whether the data variable were really of the class ‘dataset‘. Packing and unpacking of the data however causes a lot of overhead and sometimes it is entirely unnecessary. For instance when objects have just to be classified by an already trained classifier or in case of cluster analysis, it can be much more logically to stick with arrays of doubles.
Consequently, it has been decided last year that on all places where applicable doubles have to be accepted. It makes the code faster and enlarges the applicability. So calls like R*pca([],2) and R*fisherc(A) are now accepted in case R is not a dataset but just variable of the class ‘double‘. We think that we have covered all places, but one is never sure. Users who encounter suspicious error messages are encouraged to report this to us.
The applicability of PRTools is increased by this as many PRTools mappings accept now doubles. See below for an interesting example, where the new routine out2 is discussed.
Dyadic operations for mappings
Operations between variables of the PRTools programming class ‘mapping’ are originally overloaded for multiplication and concatenation. There have not been many demands to extend this. However, in order to construct a full set of operations on mappings they are now made available. They are all based on the concept that a dyadic operation between mappings should construct a new mapping, such that if it is applied to a dataset the result is identical to the same operation applied to the result of the individual mappings of that dataset. Some more information in the PRTools guide.
Here is a small example. In the figure the estimated Gaussian density (in blue) is compared with the Parzen density (in red). The black curve shows where the estimates are equal. In the center and in the outskirts is the Gaussian larger. In between, in the cluster centers wins the Parzen estimator. Look in the code and note that there the comparison of two mappings (w1 > w2) is trained.
|
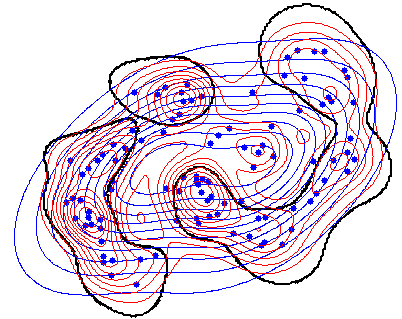 |
Note that a*(w>v) refers to two untrained mappings and results in a*w > a*v. This is again a dyadic operation, now between two trained mappings. It is applied to the plotting grid, which finally results in grid*(a*w) > grid*(a*v), which is a dyadic operation between datasets.
Programming of mappings and classifiers
For many users it appears to be difficult to program a PRTools compatible mapping or classifier. In order to make it somewhat more feasible some additional tools have been constructed:
setdefaults, handles all empty and missing input parameters in a single call.mapping_task, determines the task: mapping definition, training or execution.trained_mapping, definition of a trained mappingtrained_classifier, definition of a trained classifier
For an example, see the source code of randomforestc.
Batch processing
In an earlier version batch processing was introduced for handling large datasets by mappings. In order to avoid the construction of large intermediate arrays batches of objects are thereby executed sequentially. Originally, this was introduced as a global setting that could be switched off in case the splitting of datasets was undesirable. Users were not always aware of the problems caused by batch processing. Therefor it was decided to change this into an option that is switched off by default and can be switched on for specific mappings and classifiers that need it. There is a more extensive introduction on this topic.
Mapping output parameters
In case mappings return more than a single output parameter, these may now be retrieved when applied to a dataset by the *-operator, e.g.:
[labels,A] = kmeans(A,5);
returns the cluster indices in labels and assigns them to the dataset A as well. It can now also be written as:
[labels,A] = A*kmeans([],5);
which has the advantage that a variable W = kmeans([],5) can be supplied to another function. A new function, out2, is now available that take care that just the second output parameter is returned:
A = A*(kmeans([],5)*out2);
The brackets are needed as out2 should operate on the mapping before it is applied to the dataset. In combination with the above discussed new possibility to apply mappings to doubles, new functions can be created, e.g. the argmin function:
argmin = filtm([],'min',{[],2})*out2;
which returns in a column vector the indices of the row minima when applied to a matrix of doubles:
J = R*argmin;
as the Matlab min function returns the indices in the second parameters.
Small stuff
A routine prglobal lists and may change all global settings that determine the behaviour of PRTools
The retrieval of a single class from a dataset by the seldat command is not very straightforward. A new command, especially useful in multi-labeling problems, has been made available: selclass.
A number of classifiers shows in present version of PRTools by default a slightly different, shorter name. These are the classifier names that are stored in the mapping by setname and that can be changed by it. These names are printed during annotation of plots and on the command line. Command names have not been changed.
Filed under: PRTools

